We seek to understand the processes that modulate regulated expression of genes in cells by investigating the spatio-temporal regulation of RNA metabolism, combining single molecule and super-resolution microscopy approaches with genetics and biochemistry. Among our current research projects, we investigate how the nuclear pore complex regulates RNA transport to the cytoplasm, and how the organization of mRNP and lncRNP complexes modulates their function.
Studying the role of mRNP organization in regulating translation and mRNA metabolism
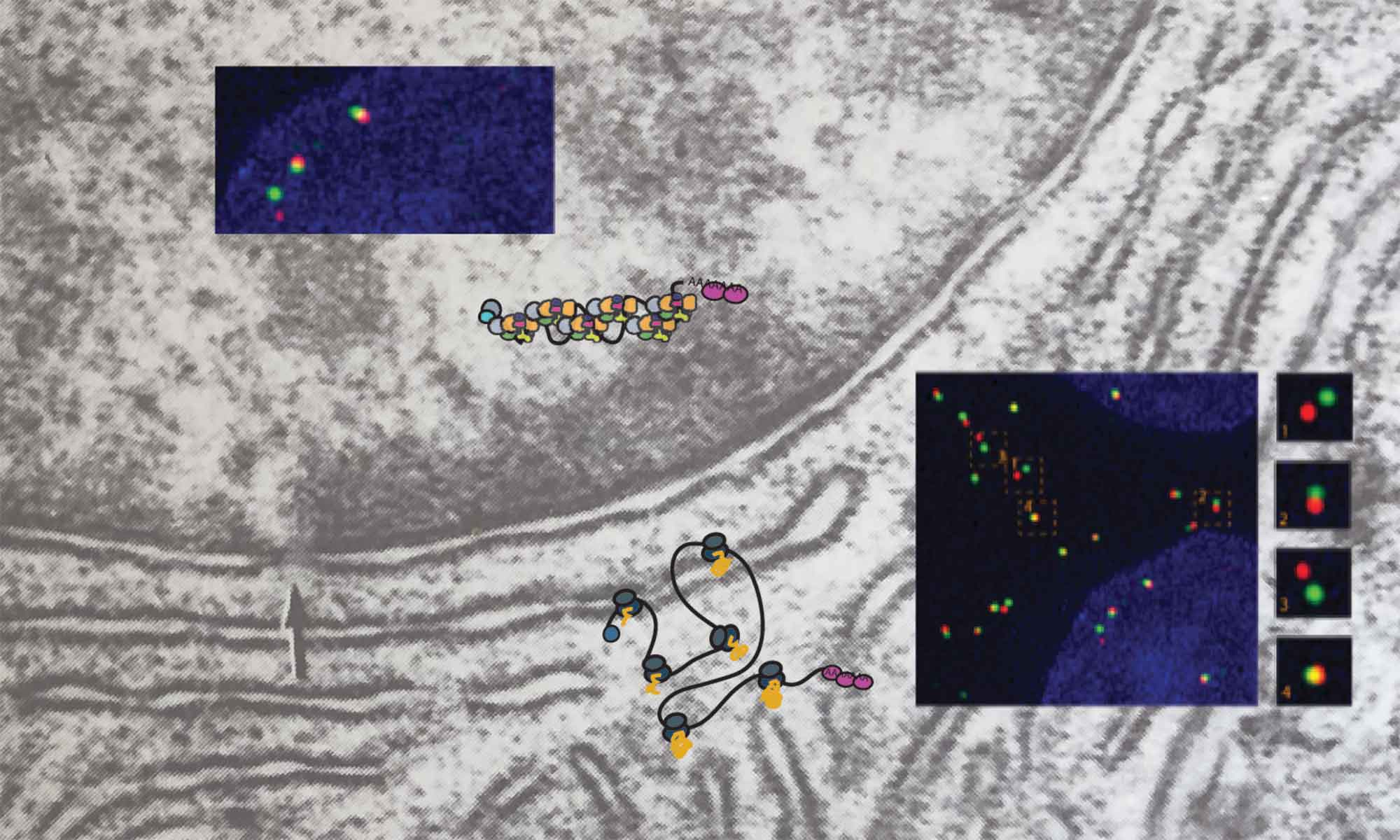
mRNAs are long nucleic acid polymers that get bound by proteins to form ribonucleoprotein complexes (mRNPs). While the mRNP proteome is well characterized, little is known about the organization of mRNP as 3D assemblies. 3D organization however is critical for regulating many aspects of RNA metabolism, such as translation, transport or degradation. We use single molecule approaches to study mRNA organization in cells and unravel its role in regulating different aspects of mRNA metabolism.
Key papers:
Adivarahan S, Livingston N, Nicholson B, Rahman S, Wu B, Rissland O, Zenklusen D. Spatial organization of single mRNPs at different stages of the gene expression pathway. Molecular Cell Published: November 8, 2018.
The nuclear pore complex and its role in mRNA export and retention
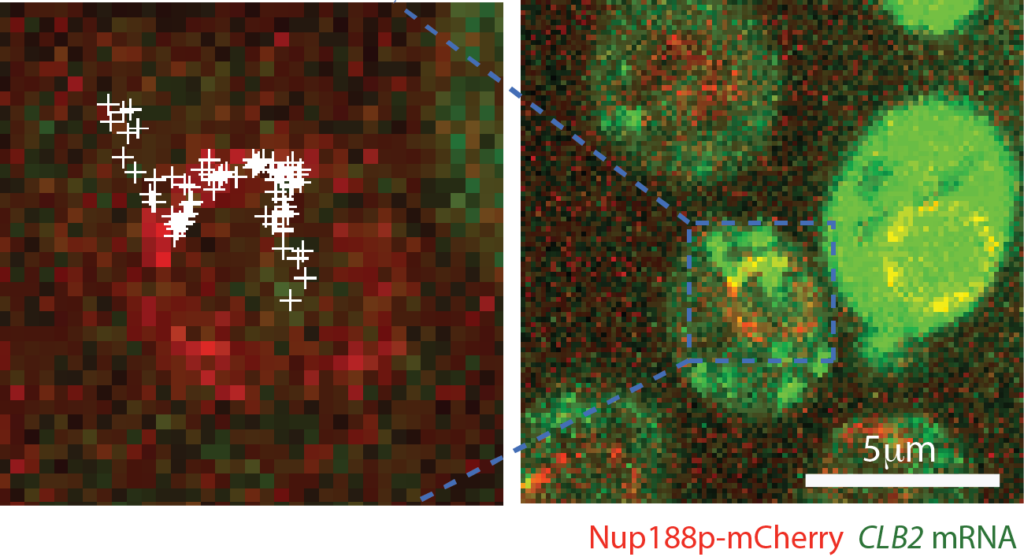
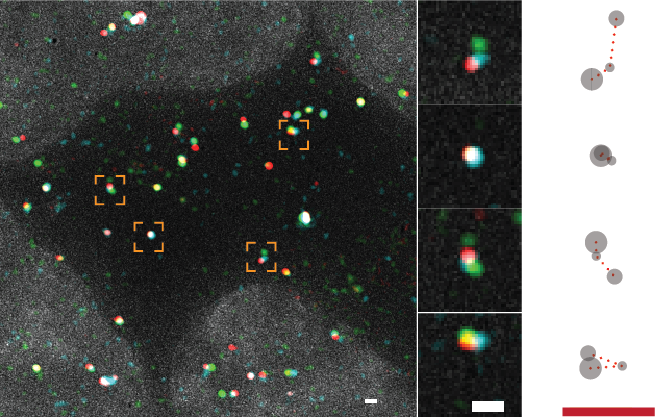
We study the mechanisms of mRNA transport from the nucleus to the cytoplasm, and the role of the nuclear pore complex (NPC) in this process. Transport to the cytoplasm is essential for mRNAs to meet with ribosomes for protein synthesis. On the other hand, many other RNA classes have to be retained in the nucleus, such us various long-non coding RNAs (lncRNAs) or mRNAs that have not been fully processed. The rules that define who is exported and who stays in the nucleus are not clear. Furthermore, many of the protein implicated in the export or in retention are found mutated in various diseases, such as ALS. Our projects investigate the relationship between the nuclear pore complex and RNA binding proteins in regulating export and retention using yeast and higher eukaryotes as model systems.
Key papers:
Bensidoun P, Reiter T, Montpetit B, Zenklusen D, Oeffinger M. Nuclear mRNA metabolism drives selective basket assembly on a subset of nuclear pore complexes in budding yeast. Molecular Cell. 2022 Oct 20;82(20):3856-3871.e6.
Fernandez-Martinez J, Kim SJ, Shi Y, Upla P, Pellarin R, Gagnon M, Chemmama IE, Wang J, Nudelman I, Zhang W, Williams R, Rice WJ, Stokes DL, Zenklusen D, Chait BT, Sali A, Rout MP. Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform. Cell 2016 Nov 17;167(5):1215-1228.e25
Saroufim MA, Bensidoun P, Raymond P, Rahman S, Krause MR, Oeffinger M, Zenklusen D. The nuclear basket mediates perinuclear mRNA scanning in budding yeast. Journal of Cell Biology 2015 Dec 21;211(6):1131-40.
Role of ncRNAs in transcription regulation
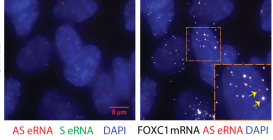
Non-coding RNA have been implicated in many aspects of transcription regulation. We are studying the role of different classes of ncRNAs in this process, including anti-sense transcription and RNAs expressed from enhancer regions (eRNAs).
Key papers:
Rahman S, Zorca C, Traboulsi T, Noutahi E, Krause M, Mader S and Zenklusen D. Single-cell profiling reveals that eRNA accumulation at enhancer-promoter loops is not required to sustain transcription. Nucleic Acid Research 2017, Apr 7;45(6):3017-3030. link
Castelnuovo M, Rahman S, Guffanti E, Infantino V, Stutz F and Zenklusen D. Bimodal expression of PHO84 is modulated by early termination of antisense transcription. Nat Struct Mol Biol. 2013 Jul;20(7):851-8. link