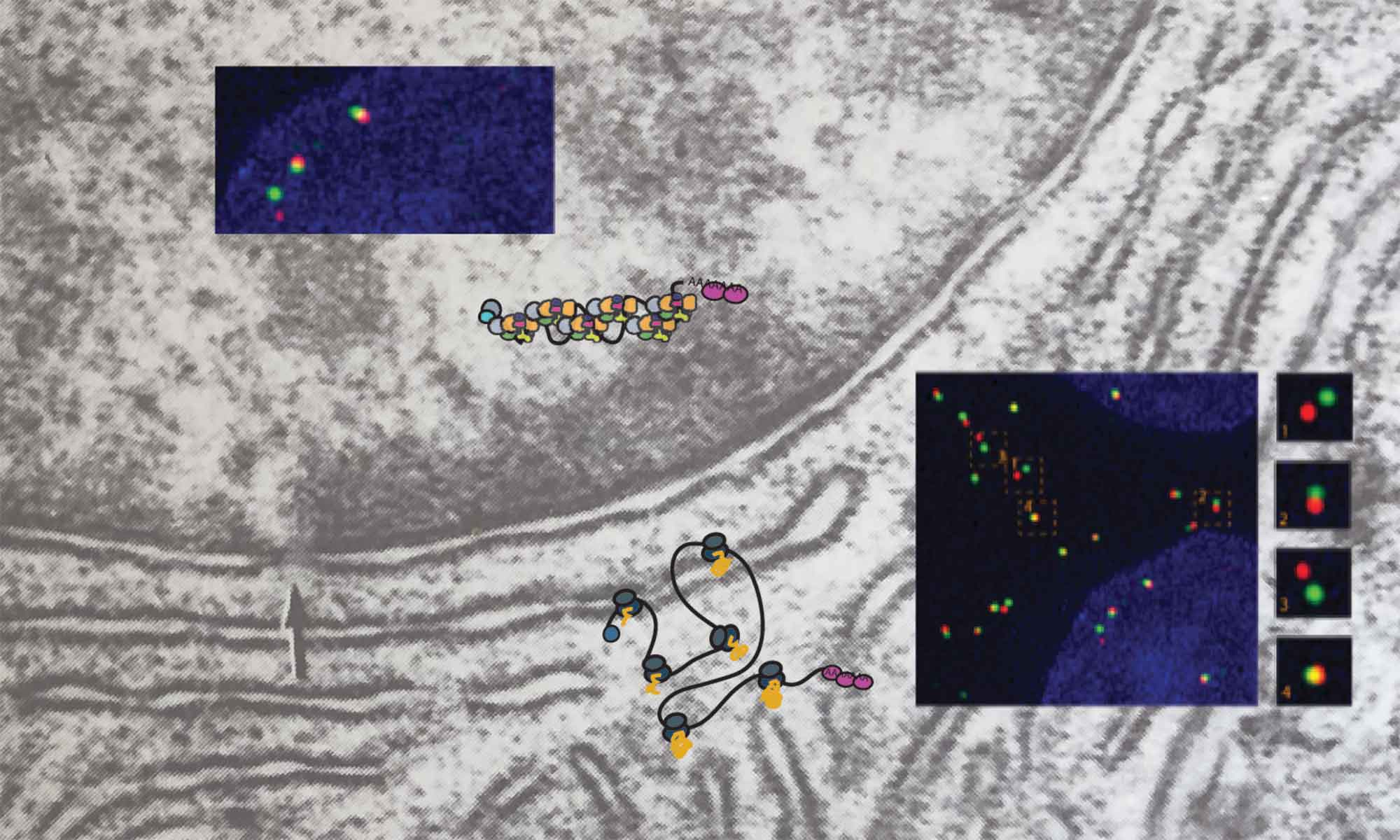
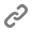Regulating gene expression, the process of reading and interpreting genetic information is a central task for all living organisms, and errors in this process cause many diseases, including cancer and various neurodegenerative diseases. We seek to understand the processes that modulate regulated expression of genes in cells by investigating the spatio-temporal regulation of RNA metabolism. Combining single molecule and super-resolution microscopy approaches with genetics and biochemistry we aim to gain a better understanding of the general rules that governing RNA metabolism, establishing the basis for a better understanding and treatment of complex diseases. Among our current research projects, we investigate how the nuclear pore complex regulates RNA transport and how the organization of mRNP and lncRNP complexes modulates their function .
We are member of the department of Biochemistry and Molecular Medicine at the Université de Montréal. Our research is supported by Canadian Institutes of Health Research, Natural Sciences and Engineering Research Council of Canada , Fonds de recherche du Québec – Santé and the Canada Foundation for Innovation.